Online game helps researchers create treatment for agricultural toxin
October 25, 2017
Researchers are using a crowdsourcing video game to face a poisonous substance that cannot be treated in a cost-effective manner.
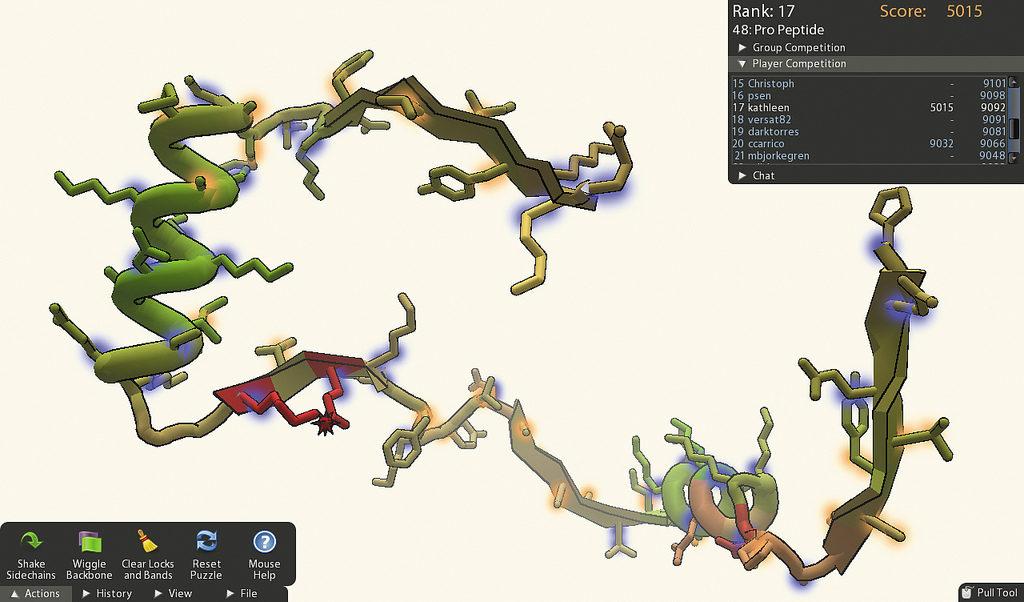
Foldit is a free online game run by Northeastern University and the University of Washington, first released to the public in 2008. Players are tasked with folding proteins into the most logical positions they can think of to help scientists predict how proteins are structured in the real world, or to help them design new proteins by modifying existing structures. The game has had over 500,000 players over its lifetime.
Aflatoxins, naturally produced by fungi common in agricultural crops, are a serious problem in third-world countries. They are known to cause liver cancer and can accumulate in cow’s milk. Altogether, around 4.5 billion people are chronically exposed to aflatoxins, according to the United States Agency for International Development.
The Aflatoxin Challenge is a collaboration between Northeastern University, the University of Washington, the University of California, Davis, food manufacturer MARS Inc. and biotechnology company Thermofisher Scientific. The first puzzle for the Aflatoxin Challenge began Oct. 13and will continue until Oct. 31. Seth Cooper, an assistant professor in the College of Computer and Information Science, has been working on Foldit since development began in 2007 and wrote his Ph.D. dissertation about it.
“The game has different mechanics and rules that can be applied to parts of the protein structure — some parts are connected to other parts, some parts will be locked in place so you can’t move them, some parts are designable so you can actually change the atoms and some parts you can’t and so on,” Cooper said.
Foldit is a complex game, but a degree in biochemistry isn’t required to participate, and a majority of players have no experience in the field. The game has a series of tutorials designed to bring anyone up to speed, with the goal of allowing new players to compete with veterans as quickly as possible. Clear goals, immediate feedback, and custom timescales are all key to the onboarding process, said Cooper.
“I think that it’s a pretty good idea to have people who may not have a trained eye because I feel like this gives a larger sample size and a larger possibility for catching things that you wouldn’t have seen in the first place,” said Tom Collins, a second-year biochemistry major. “Though I feel like there could be a point where it’s kind of a burden financially.”
Feedback is given through points-based scoring. The way Foldit calculates how many points a structure is worth goes back to its origins.
“The game is structured as sort of a leaderboard competition, and the reason we did it that way is because the game is built on top of a piece of biochemistry software called Rosetta,” Cooper said. “Rosetta can look at a protein structure and basically give you a number that tells you roughly speaking how well-folded and designed the protein is.”
The puzzles, generally open for a week at a time, have players compete alone or in teams to push each other further and further up the leaderboard to achieve the most desirable result. At the end, researchers analyze the results and use them to alter the next challenge, said Cooper. Puzzles get progressively more complex as players are allowed to manipulate the proteins in more ways.
The final submission with the highest score isn’t necessarily the solution, however. The top scoring submissions are considered the most likely candidates, and multiple submissions are tested for accuracy, Cooper said. Testing for aflatoxin challengers will be done at the Siegel Lab at the University of California, Davis. Heading the lab is Justin Siegel, an assistant professor of the UC Davis department of chemistry. Siegel, like Cooper, has been working on Foldit since its inception.
“Our role going forward, now that we’ve identified the enzyme, is that we’ll be testing all of the enzymes that come from the puzzle,” Siegel said.
The enzyme in the Aflatoxin Challenge is called gluconolactanase. It can dissolve a lactone ring in aflatoxin, an action that would massively reduce the substance’s toxicity. However, the active site of the enzyme does not recognize aflatoxin. The goal of the challenge is to modify the active site to fit its new target, according to the challenge announcement.
In order to test the winning submissions, Siegel and his team have to actually make them first. This is done using a process called gene synthesis, accomplished using Thermofisher’s own proprietary method.
“We can type into the computer whatever DNA sequence we want and since we know how DNA translates into proteins, we know our protein can back-translate into DNA,” Siegel said. “We type that into the computer, the computer actually back-translates the code and then it’s just like typing it into a Word file and when you hit print, out comes the DNA.”