Northeastern researchers advise on current, future pandemics
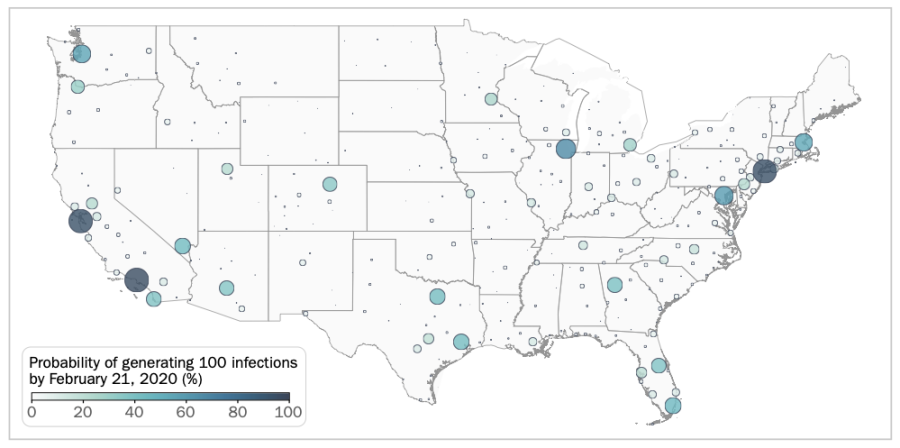
One of the MoBS lab models looked at the likelihood of different regions to spread COVID-19 internally. Image courtesy of MoBS lab, via “Cryptic transmission of SARS-CoV-2 and the first COVID-19 wave.”
December 16, 2021
The speed at which COVID-19 has spread and the length of time it has persisted highlights the lack of preparedness for handling pandemics in an increasingly interconnected world. Researchers at Northeastern’s Laboratory for the Modeling of Biological and Sociotechnical Systems, or MoBS lab, are using the spread of COVID-19 to create models of epidemiological spread that can be used to stay on top of future pandemics.
The latest COVID-19 research from the MoBS lab, led by Northeastern doctoral candidate Jessica Davis and run by Northeastern professor Alessandro Vespignani, was published online Oct. 25 as an unedited early access paper in the scientific journal Nature. It was also featured in a Boston Globe article later that day.
“The goal of this paper was to provide an anatomy of the first wave of the COVID-19 pandemic,” Davis said in an interview with The News.
This paper used a scenario modeling program called Global Epidemic and Mobility, or GLEAM, model to predict likely sites and dates of COVID-19 introduction and to trace the transmission resulting from each introduction, based on known regional travel and infection rates.
“Our model essentially takes the world and chops it up into little pieces. Within each region, we know how many people live there, and we know how many people travel between regions,” Davis said.
The MoBS researchers used the model to take a retrospective look at how COVID-19 spread during the “cryptic” period, which Vespignani describes as a phase where there was transmission, but cases went unnoticed because of limited testing.
“We tried to reconstruct what was happening during [January and February 2020], where the cases were coming from, what is the likelihood that specific urban areas had a specific number of infections, and so on and so forth,” Vespignani said.
With the model, Davis and the rest of the MoBS team predicted that COVID-19 could have been introduced to the United States as early as December 2019, with local transmission likely occurring first in California at the end of January 2020.
The first confirmed Massachusetts case, and the eighth U.S. case, was identified Feb. 1, 2020. The model predicted that by April 2020, domestic transmission was responsible for between 60%-80% of viral introductions in Massachusetts.
“Part of our work was, if we were to simulate this epidemic over and over again, what are the likely sources of importation to different U.S. states and different European countries?” Davis said.
These simulations were combined to predict the most likely sources of importation and local transmission during the time period with limited testing.
“This is absolutely beautiful work by Alessandro Vespignani and colleagues, rigorously examining the phenomena of hidden transmission of infectious diseases,” wrote Pardis Sabeti, a Harvard professor and principal investigator of the Sabeti Lab, in an email statement to The News.
Davis and Vespignani’s work on this paper began in February 2020, using modeling systems developed from the MoBS lab’s prior research on seasonal influenza, the 2009 swine flu pandemic, the 2013 Ebola outbreak and the 2015 Zika epidemic. Their early work included consulting with agencies looking for the best way to handle the pandemic.
“We’ve been working with the World Health Organization [WHO] and the Centers for Disease Control and Prevention [CDC], and our team was part of the modeling teams that were advising the White House task force,” Vespignani said.
The model highlights the importance of immediate proactive prevention, such as increased testing, rather than delayed reactive responses, like travel restrictions. The actual spread, which was mainly controlled by reactive responses, indicates the truth in this finding.
“What we propose in the paper is that, in the future, we need not to play these catch up games in which we say, ‘Well, there are cases in China,’ and then we test people from China,” Vespignani said. “When you see that there is already an outbreak in a country, you probably already have outbreaks in other places, so you have to be more comprehensive in your testing policies.”
“With how connected we are around the world, it is of the utmost importance to better understand cryptic transmission during a pandemic,” Sabeti said in her statement.
The MoBS lab plans to use this data to refine their models. However, they hope that a refined model could be able to predict and prevent further COVID-19 spread by testing the effects of possible variants and situation changes.
“We can look at, for example, what would happen if we started introducing childhood vaccination, or what would happen if there was another variant that was more transmissible than [the Delta variant],” Davis said.
Beyond COVID-19, the model shows promise for predicting the spread of any pandemic.
“The kinds of modeling approaches that we define in this paper and that we have used in other situations can provide a guide to the policy makers to say, ‘Here’s where it can come from, where we should focus our attention, how we should prepare,’” Vespignani said. “We can use those tools not as a response tool on the field, but as an intelligence information that you can use to optimize your approaches to mitigation and containment.”
The MoBS lab continues to consult with several health agencies, including the CDC and WHO, in collaboration with epidemiology labs across the country, to control the spread of COVID-19 variants and help the regions that continue to experience surges of pandemic activity.